Download the compressed program archive
On the SARST2 download page, click on the compressed file that matches your operating system and CPU architecture to download it.
How to know your CPU architecture?
Windows
- Method 1:
Start button (Windows icon) > Settings > System > About > Device specifications > System type
"x64_based processor" means x86_64 architecture
"ARM_based processor" means ARM64 architecture - Method 2:
Open Command Prompt. Type the following command, then press Enter:echo %PROCESSOR_ARCHITECTURE%
If you get
AMD64
please download "x86_64" version
ARM64
please download "ARM64" version
Linux/macOS
- Open Terminal
 and type the following command, then press Enter:
and type the following command, then press Enter:uname -m
If you get
x86_64
please download "x86_64" version
aarch64
please download "ARM64" version
How to Open Windows Command Prompt (CMD)?
Click the Start button and type "cmd" into the search bar. Click on "Command Prompt" in the search results.
Decompress the archive
Windows
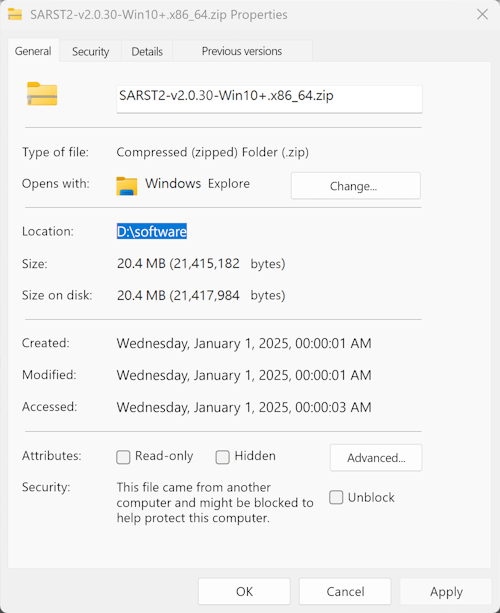
- Right-click the compressed file, then go to Properties > General > Location to find the file's path (for example, "D:\software").
- Open Windows Command Prompt (CMD) and change to the drive where your file is located.
C:\User>D:
- Change to the directory where your file is located.
D:\>cd D:\software
- Extract the compressed file.
- x86_64
- zip file
tar -xf SARST2-v2.0.30-Win10+.x86_64.zip
- tar.gz file
- ARM64
- zip file
- tar.gz file
tar -zxvf SARST2-v2.0.30-Win10+.x86_64.tar.gz
tar -xf SARST2-v2.0.30-Win10+.arm64.zip
tar -zxvf SARST2-v2.0.30-Win10+.arm64.tar.gz
Linux/macOS
Open Terminal![]() , navigate to the directory where your compressed file is located. Then, type the following command (using Linux as an example) and press Enter:
, navigate to the directory where your compressed file is located. Then, type the following command (using Linux as an example) and press Enter:
- x86_64
- zip file
unzip SARST2-v2.0.30-Linux.x86_64.zip
tar -xzf SARST2-v2.0.30-Linux.x86_64.tar.gz
- zip file
unzip SARST2-v2.0.30-Linux.arm64.zip
tar -xzf SARST2-v2.0.30-Linux.arm64.tar.gz
Run the program to show the usage
Windows
Navigate to the directory where your program is located.
cd SARST2-v2.0.30-Win10+.x86_64\binRunning any of the above programs without parameters (or with -h) will display a brief text-based help message for that program.
sarst2
.\sarst2.exeformatdb
.\formatdb.exereaddb
.\readdb.exeLinux/macOS
Navigate to the directory where your program is located (using Linux as an example).
cd SARST2-v2.0.30-Linux.x86_64/binRunning any of the above programs without parameters (or with -h) will display a brief text-based help message for that program.
sarst2
./sarst2formatdb
./formatdbreaddb
./readdbExample Usage
sarst2
sarst2
Pairwise alignment
on Windows
.\sarst2.exe ..\dat\1atp.pdb ..\dat\5uzkA.pdbon Linux/macOS
./sarst2 ../dat/1atp.pdb ../dat/5uzkA.pdbExample result
D:\software>cd SARST2-v2.0.30-Win10+.x86_64\bin
D:\software\SARST2-v2.0.30-Win10+.x86_64\bin>.\sarst2.exe ..\dat\1atp.pdb ..\dat\5uzkA.pdb
+------------------------------------------------------------------------------+
| SARST (v2.0.30, build 20250609) |
| Structural similarity search Aided by Ramachandran Sequential Transformation |
| |
| Authors: Wei-Cheng Lo, Arieh Warshel, Chia-Hua Lo, et al. |
| Article: Nature Communications, 2025, DOI: 10.1038/s41467-025-63757-9 |
+------------------------------------------------------------------------------+
## Query: ..\dat\1atp.pdb
Protein length = 336 residues
## Sbjct: ..\dat\5uzkA.pdb
Protein length = 337 residues
Aligned residues = 336 residues
RMSD = 0.5557 Å
Seq identity = 97.92 % (329/336)
Seq similarity = 99.70 % (335/336)
Confidence-score = 0.9795
pC-value = 0.0299
TM-score, nQuery = 0.9936 (norm size: 336 res, d0: 6.69 Å)
Query: 1 -VKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMKI 59
VKEFLAKAKEDFLKKWE+P+QNTA:LDQF+RIKTLGTGSFGRVMLVKHKE+GNHYAMKI
Sbjct: 1 SVKEFLAKAKEDFLKKWESPAQNTAHLDQFERIKTLGTGSFGRVMLVKHKETGNHYAMKI 60
Query: 60 LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHLR 119
LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYV.GGEMFSHLR
Sbjct: 61 LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVPGGEMFSHLR 120
Query: 120 RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG 179
RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG
Sbjct: 121 RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG 180
Query: 180 RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG 239
RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG
Sbjct: 181 RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG 240
Query: 240 KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP 299
KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP
Sbjct: 241 KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP 300
Query: 300 FIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF 336
FIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEF+EF
Sbjct: 301 FIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFSEF 337One-against-all alignment
on Windows
.\sarst2.exe ..\dat\1atp.pdb ..\dat\5uzkA.pdb ..\dat\6p5pC.pdb ..\dat\101m.pdbon Linux/macOS
./sarst2 ../dat/1atp.pdb ../dat/5uzkA.pdb ../dat/6p5pC.pdb ../dat/101m.pdbExample result
D:\software>cd SARST2-v2.0.30-Win10+.x86_64\bin
D:\software\SARST2-v2.0.30-Win10+.x86_64\bin>.\sarst2.exe ..\dat\1atp.pdb ..\dat\5uzkA.pdb ..\dat\6p5pC.pdb ..\dat\101m.pdb
+------------------------------------------------------------------------------+
| SARST (v2.0.30, build 20250609) |
| Structural similarity search Aided by Ramachandran Sequential Transformation |
| |
| Authors: Wei-Cheng Lo, Arieh Warshel, Chia-Hua Lo, et al. |
| Article: Nature Communications, 2025, DOI: 10.1038/s41467-025-63757-9 |
+------------------------------------------------------------------------------+
3
## Query: ..\dat\1atp.pdb
Protein size = 336 residues
Subjects:
Dataset size = 3 structure(s)
..\dat\5uzkA.pdb, ..\dat\6p5pC.pdb, ..\dat\101m.pdb
================================================================================
Ranking Protein Size Ident(%) ASize RMSD(Å) TM-score Conf
--------------------------------------------------------------------------------
1 5uzkA 337 97.92 336 0.5557 0.9936 0.9822
2 6p5pC 375 32.74 299 2.1978 0.8291 0.7904
================================================================================
>> 1
## Sbjct: ..\dat\5uzkA.pdb
Protein length = 337 residues
Aligned residues = 336 residues
RMSD = 0.5557 Å
Seq identity = 97.92 % (329/336)
Seq similarity = 99.70 % (335/336)
Confidence-score = 0.9822
pC-value = 0.0259
TM-score, nQuery = 0.9936 (norm size: 336 res, d0: 6.69 Å)
Query: 1 -VKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMKI 59
VKEFLAKAKEDFLKKWE+P+QNTA:LDQF+RIKTLGTGSFGRVMLVKHKE+GNHYAMKI
Sbjct: 1 SVKEFLAKAKEDFLKKWESPAQNTAHLDQFERIKTLGTGSFGRVMLVKHKETGNHYAMKI 60
Query: 60 LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHLR 119
LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYV.GGEMFSHLR
Sbjct: 61 LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVPGGEMFSHLR 120
Query: 120 RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG 179
RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG
Sbjct: 121 RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG 180
Query: 180 RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG 239
RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG
Sbjct: 181 RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG 240
Query: 240 KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP 299
KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP
Sbjct: 241 KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP 300
Query: 300 FIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF 336
FIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEF+EF
Sbjct: 301 FIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFSEF 337
>> 2
## Sbjct: ..\dat\6p5pC.pdb
Protein length = 375 residues
Aligned residues = 299 residues
RMSD = 2.1978 Å
Seq identity = 32.74 % (110/336)
Seq similarity = 54.76 % (184/336)
Confidence-score = 0.7904
pC-value = 0.3393
TM-score, nQuery = 0.8291 (norm size: 336 res, d0: 6.69 Å)
Query: 1 VKEFLAKAKEDFLKKWETPSQ--------------------------------------- 21
Sbjct: 1 ---------------------RKLEALIRDPRSPINVESLLDGLNSLVLDLDFPALRKNK 39
Query: 22 ------------------NTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMKILDKQ 63
...+.+.+D.+K.+G.G+FG.V.LV+HK.S...YAMK+L.K.
Sbjct: 40 NIDNFLNRYEKIVKKIRGLQMKAEDYDVVKVIGRGAFGEVQLVRHKASQKVYAMKLLSKF 99
Query: 64 KVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHLRRIGR 123
::+K.........E+.I+...N.P::V+L.++F+D+..LYMVMEY:.GG++.:.+. ...
Sbjct: 100 EMIKRSDSAFFWEERDIMAFANSPWVVQLFYAFQDDRYLYMVMEYMPGGDLVNLMS-NYD 158
Query: 124 FSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG---- 179
..E..A+FY.A++VL.:+.+HS+.LI+RD:KP+N+L+D+:G+++:.DFG...+:.
Sbjct: 159 VPEKWAKFYTAEVVLALDAIHSMGLIHRDVKPDNMLLDKHGHLKLADFGTCMKMD-ETGM 217
Query: 180 -RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVS 238
......GTP+Y++PE++.S+ Y.+..DWW+:GV:+YEM..G..PF+AD..+..Y.KI:.
Sbjct: 218 VHCDTAVGTPDYISPEVLKSQ-YGRECDWWSVGVFLYEMLVGDTPFYADSLVGTYSKIMD 276
Query: 239 GKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEA 298
..:.FP+..S...K+L+..:L. D...R: .+NGV.+I+.H.:F....W.....R:..A
Sbjct: 277 HSLCFPAEISKHAKNLICAFLT-DREVRL--GRNGVEEIRQHPFFKNDQWHWDNIRETAA 333
Query: 299 PFIPKFKGPGDTSNFDDYEEEEIRVXIN-----------EKCGKE--FTEF- 336
P.+P::....D+SNFDD.E ...... FT.
Sbjct: 334 PVVPELSSDIDSSNFDDIE---------VETFPIPKAFVGNQLPFIGFTY-Y 375Database searching (a database formatted by formatdb is required)
on Windows
.\sarst2.exe ..\dat\1atp.pdb -db mySarstDbon Linux/macOS
./sarst2 ../dat/1atp.pdb -db mySarstDbExample result
D:\software>cd SARST2-v2.0.30-Win10+.x86_64\bin
D:\software\SARST2-v2.0.30-Win10+.x86_64\bin>.\sarst2.exe ..\dat\1atp.pdb -db mySarstDb
+------------------------------------------------------------------------------+
| SARST (v2.0.30, build 20250609) |
| Structural similarity search Aided by Ramachandran Sequential Transformation |
| |
| Authors: Wei-Cheng Lo, Arieh Warshel, Chia-Hua Lo, et al. |
| Article: Nature Communications, 2025, DOI: 10.1038/s41467-025-63757-9 |
+------------------------------------------------------------------------------+
## Query: ..\dat\1atp.pdb
Protein size = 336 residues
Subjects:
Dataset size = 100 structure(s)
Database = mySarstDb
================================================================================
Ranking Protein Size Ident(%) ASize RMSD(Å) TM-score Conf
--------------------------------------------------------------------------------
1 1atpE 336 100.00 336 0.0000 1.0000 0.9957
2 2ojfE 336 97.02 336 0.5339 0.9941 0.9838
3 2gfcA 337 97.62 336 0.5144 0.9946 0.9821
4 2oh0E 336 97.02 336 0.5963 0.9926 0.9813
5 1q61A 337 96.73 336 0.5151 0.9942 0.9812
6 3p0mA 337 97.92 336 0.5570 0.9934 0.9809
7 1sveA 344 97.02 334 0.5245 0.9883 0.9772
8 1xh9A 338 96.13 336 0.7475 0.9889 0.9767
9 2gniA 341 95.54 336 0.8804 0.9894 0.9753
10 3l9lA 340 97.32 336 0.9409 0.9830 0.9737
11 1q8wA 337 97.32 336 0.9435 0.9813 0.9735
12 1rekA 338 100.00 336 1.0469 0.9805 0.9735
13 1xhaA 350 95.83 334 0.7475 0.9823 0.9695
14 3l9nA 334 95.83 333 1.0637 0.9717 0.9670
15 1cmkE 350 96.43 335 2.0660 0.9303 0.9453
16 1sykA 350 98.51 332 2.7546 0.8821 0.9213
17 3ag9A 325 93.15 321 2.5384 0.8580 0.9096
18 4dfyA 311 91.07 306 3.2413 0.7769 0.8600
19 3e8dA 316 38.10 298 1.3609 0.8577 0.8123
20 7atvA 328 17.86 257 3.1805 0.6660 0.6647
21 3at2A 334 16.67 239 2.8027 0.6360 0.6530
================================================================================
>> 1
## Sbjct: ..\dat\PDB-sample\1atpE.pdb
Protein length = 336 residues
Aligned residues = 336 residues
RMSD = 0.0000 Å
Seq identity = 100.00 % (336/336)
Seq similarity = 100.00 % (336/336)
Confidence-score = 0.9957
pC-value = 0.0062
TM-score, nQuery = 1.0000 (norm size: 336 res, d0: 6.69 Å)
Query: 1 VKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMKIL 60
VKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMKIL
Sbjct: 1 VKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMKIL 60
Query: 61 DKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHLRR 120
DKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHLRR
Sbjct: 61 DKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHLRR 120
Query: 121 IGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKGR 180
IGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKGR
Sbjct: 121 IGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKGR 180
Query: 181 TWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSGK 240
TWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSGK
Sbjct: 181 TWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSGK 240
Query: 241 VRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAPF 300
VRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAPF
Sbjct: 241 VRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAPF 300
Query: 301 IPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF 336
IPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF
Sbjct: 301 IPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF 336
>> 2
## Sbjct: ..\dat\PDB-sample\2ojfE.pdb
Protein length = 336 residues
Aligned residues = 336 residues
RMSD = 0.5339 Å
Seq identity = 97.02 % (326/336)
Seq similarity = 98.51 % (331/336)
Confidence-score = 0.9838
pC-value = 0.0236
TM-score, nQuery = 0.9941 (norm size: 336 res, d0: 6.69 Å)
Query: 1 VKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMKIL 60
VKEFLAKAKEDFLKKWE.P+QNTA:LDQF+RIKTLGTGSFGRVMLVKH.E+GNHYAMKIL
Sbjct: 1 VKEFLAKAKEDFLKKWENPAQNTAHLDQFERIKTLGTGSFGRVMLVKHMETGNHYAMKIL 60
Query: 61 DKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHLRR 120
DKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYV.GGEMFSHLRR
Sbjct: 61 DKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVPGGEMFSHLRR 120
Query: 121 IGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKGR 180
IGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKGR
Sbjct: 121 IGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKGR 180
Query: 181 TWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSGK 240
TW.LCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSGK
Sbjct: 181 TWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSGK 240
Query: 241 VRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAPF 300
VRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAPF
Sbjct: 241 VRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAPF 300
Query: 301 IPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF 336
IPKFKGPGDTSNFDDYEEEEIRV.INEKCGKEF+EF
Sbjct: 301 IPKFKGPGDTSNFDDYEEEEIRVSINEKCGKEFSEF 336
>> 3
## Sbjct: ..\dat\PDB-sample\2gfcA.pdb
Protein length = 337 residues
Aligned residues = 336 residues
RMSD = 0.5144 Å
Seq identity = 97.62 % (328/336)
Seq similarity = 99.11 % (333/336)
Confidence-score = 0.9821
pC-value = 0.0260
TM-score, nQuery = 0.9946 (norm size: 336 res, d0: 6.69 Å)
Query: 1 -VKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMKI 59
VKEFLAKAKEDFLKKWE.P+QNTA:LDQF+RIKTLGTGSFGRVMLVKH.E+GNHYAMKI
Sbjct: 1 SVKEFLAKAKEDFLKKWENPAQNTAHLDQFERIKTLGTGSFGRVMLVKHMETGNHYAMKI 60
Query: 60 LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHLR 119
LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYV.GGEMFSHLR
Sbjct: 61 LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVPGGEMFSHLR 120
Query: 120 RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG 179
RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG
Sbjct: 121 RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG 180
Query: 180 RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG 239
RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG
Sbjct: 181 RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG 240
Query: 240 KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP 299
KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP
Sbjct: 241 KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP 300
Query: 300 FIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF 336
FIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEF+EF
Sbjct: 301 FIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFSEF 337
>> 4
## Sbjct: ..\dat\PDB-sample\2oh0E.pdb
Protein length = 336 residues
Aligned residues = 336 residues
RMSD = 0.5963 Å
Seq identity = 97.02 % (326/336)
Seq similarity = 98.51 % (331/336)
Confidence-score = 0.9813
pC-value = 0.0272
TM-score, nQuery = 0.9926 (norm size: 336 res, d0: 6.69 Å)
Query: 1 VKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMKIL 60
VKEFLAKAKEDFLKKWE.P+QNTA:LDQF+RIKTLGTGSFGRVMLVKH.E+GNHYAMKIL
Sbjct: 1 VKEFLAKAKEDFLKKWENPAQNTAHLDQFERIKTLGTGSFGRVMLVKHMETGNHYAMKIL 60
Query: 61 DKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHLRR 120
DKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYV.GGEMFSHLRR
Sbjct: 61 DKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVPGGEMFSHLRR 120
Query: 121 IGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKGR 180
IGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKGR
Sbjct: 121 IGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKGR 180
Query: 181 TWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSGK 240
TW.LCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSGK
Sbjct: 181 TWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSGK 240
Query: 241 VRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAPF 300
VRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAPF
Sbjct: 241 VRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAPF 300
Query: 301 IPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF 336
IPKFKGPGDTSNFDDYEEEEIRV.INEKCGKEF+EF
Sbjct: 301 IPKFKGPGDTSNFDDYEEEEIRVSINEKCGKEFSEF 336
>> 5
## Sbjct: ..\dat\PDB-sample\1q61A.pdb
Protein length = 337 residues
Aligned residues = 336 residues
RMSD = 0.5151 Å
Seq identity = 96.73 % (325/336)
Seq similarity = 98.81 % (332/336)
Confidence-score = 0.9812
pC-value = 0.0273
TM-score, nQuery = 0.9942 (norm size: 336 res, d0: 6.69 Å)
Query: 1 -VKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMKI 59
VKEFLAKAKEDFLKKWE.P+QNTA:LDQF+RIKTLGTGSFGRVMLVKH.E+GNHYAMKI
Sbjct: 1 SVKEFLAKAKEDFLKKWENPAQNTAHLDQFERIKTLGTGSFGRVMLVKHMETGNHYAMKI 60
Query: 60 LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHLR 119
LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEY..GGEMFSHLR
Sbjct: 61 LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYAPGGEMFSHLR 120
Query: 120 RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG 179
RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENL+IDQQGYI+VTDFGFAKRVKG
Sbjct: 121 RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLMIDQQGYIKVTDFGFAKRVKG 180
Query: 180 RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG 239
RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG
Sbjct: 181 RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG 240
Query: 240 KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP 299
KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP
Sbjct: 241 KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP 300
Query: 300 FIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF 336
FIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEF+EF
Sbjct: 301 FIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFSEF 337
>> 6
## Sbjct: ..\dat\PDB-sample\3p0mA.pdb
Protein length = 337 residues
Aligned residues = 336 residues
RMSD = 0.5570 Å
Seq identity = 97.92 % (329/336)
Seq similarity = 99.70 % (335/336)
Confidence-score = 0.9809
pC-value = 0.0278
TM-score, nQuery = 0.9934 (norm size: 336 res, d0: 6.69 Å)
Query: 1 -VKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMKI 59
VKEFLAKAKEDFLKKWE+P+QNTA:LDQF+RIKTLGTGSFGRVMLVKHKE+GNHYAMKI
Sbjct: 1 SVKEFLAKAKEDFLKKWESPAQNTAHLDQFERIKTLGTGSFGRVMLVKHKETGNHYAMKI 60
Query: 60 LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHLR 119
LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYV.GGEMFSHLR
Sbjct: 61 LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVPGGEMFSHLR 120
Query: 120 RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG 179
RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG
Sbjct: 121 RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG 180
Query: 180 RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG 239
RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG
Sbjct: 181 RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG 240
Query: 240 KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP 299
KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP
Sbjct: 241 KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP 300
Query: 300 FIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF 336
FIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEF+EF
Sbjct: 301 FIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFSEF 337
>> 7
## Sbjct: ..\dat\PDB-sample\1sveA.pdb
Protein length = 344 residues
Aligned residues = 334 residues
RMSD = 0.5245 Å
Seq identity = 97.02 % (326/336)
Seq similarity = 98.51 % (331/336)
Confidence-score = 0.9772
pC-value = 0.0333
TM-score, nQuery = 0.9883 (norm size: 336 res, d0: 6.69 Å)
Query: 1 VK----------EFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKE 50
EFLAKAKEDFLKKWE.P+QNTA:LDQF+RIKTLGTGSFGRVMLVKH.E
Sbjct: 1 --KKGXEQESVKEFLAKAKEDFLKKWENPAQNTAHLDQFERIKTLGTGSFGRVMLVKHME 58
Query: 51 SGNHYAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVA 110
+GNHYAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYV.
Sbjct: 59 TGNHYAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVP 118
Query: 111 GGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTD 170
GGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTD
Sbjct: 119 GGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTD 178
Query: 171 FGFAKRVKGRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPI 230
FGFAKRVKGRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPI
Sbjct: 179 FGFAKRVKGRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPI 238
Query: 231 QIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIA 290
QIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIA
Sbjct: 239 QIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIA 298
Query: 291 IYQRKVEAPFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF 336
IYQRKVEAPFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEF+EF
Sbjct: 299 IYQRKVEAPFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFSEF 344
>> 8
## Sbjct: ..\dat\PDB-sample\1xh9A.pdb
Protein length = 338 residues
Aligned residues = 336 residues
RMSD = 0.7475 Å
Seq identity = 96.13 % (323/336)
Seq similarity = 98.81 % (332/336)
Confidence-score = 0.9767
pC-value = 0.0340
TM-score, nQuery = 0.9889 (norm size: 336 res, d0: 6.69 Å)
Query: 1 --VKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMK 58
VKEFLAKAKEDFLKKWE.P+QNTA:LDQF+RIKTLGTGSFGRVMLVKH.E+GNHYAMK
Sbjct: 1 ESVKEFLAKAKEDFLKKWENPAQNTAHLDQFERIKTLGTGSFGRVMLVKHMETGNHYAMK 60
Query: 59 ILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHL 118
ILDKQKVVKLK+IEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEY..GGEMFSHL
Sbjct: 61 ILDKQKVVKLKEIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYAPGGEMFSHL 120
Query: 119 RRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVK 178
RRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENL+IDQQGYI+VTDFG:AKRVK
Sbjct: 121 RRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLMIDQQGYIKVTDFGLAKRVK 180
Query: 179 GRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVS 238
GRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVS
Sbjct: 181 GRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVS 240
Query: 239 GKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEA 298
GKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEA
Sbjct: 241 GKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEA 300
Query: 299 PFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF 336
PFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEF+EF
Sbjct: 301 PFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFSEF 338
>> 9
## Sbjct: ..\dat\PDB-sample\2gniA.pdb
Protein length = 341 residues
Aligned residues = 336 residues
RMSD = 0.8804 Å
Seq identity = 95.54 % (321/336)
Seq similarity = 98.51 % (331/336)
Confidence-score = 0.9753
pC-value = 0.0360
TM-score, nQuery = 0.9894 (norm size: 336 res, d0: 6.69 Å)
Query: 1 VK-----EFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHY 55
.: EFLAKAKEDFLKKWE.P+QNTA:LDQF+RIKT+GTGSFGRVMLVKH.E+GNHY
Sbjct: 1 XEQESVKEFLAKAKEDFLKKWENPAQNTAHLDQFERIKTIGTGSFGRVMLVKHMETGNHY 60
Query: 56 AMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMF 115
AMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEY:.GG+MF
Sbjct: 61 AMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYMPGGDMF 120
Query: 116 SHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAK 175
SHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYI+V.DFGFAK
Sbjct: 121 SHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIKVADFGFAK 180
Query: 176 RVKGRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEK 235
RVKGRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEK
Sbjct: 181 RVKGRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEK 240
Query: 236 IVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRK 295
IVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRK
Sbjct: 241 IVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRK 300
Query: 296 VEAPFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF 336
VEAPFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEF+EF
Sbjct: 301 VEAPFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFSEF 341
>> 10
## Sbjct: ..\dat\PDB-sample\3l9lA.pdb
Protein length = 340 residues
Aligned residues = 336 residues
RMSD = 0.9409 Å
Seq identity = 97.32 % (327/336)
Seq similarity = 99.40 % (334/336)
Confidence-score = 0.9737
pC-value = 0.0384
TM-score, nQuery = 0.9830 (norm size: 336 res, d0: 6.69 Å)
Query: 1 VK----EFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYA 56
.+ EFLAKAKEDFLKKWE+P+QNTA:LDQF+RIKTLGTGSFGRVMLVKHKE+GNHYA
Sbjct: 1 EQESVKEFLAKAKEDFLKKWESPAQNTAHLDQFERIKTLGTGSFGRVMLVKHKETGNHYA 60
Query: 57 MKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFS 116
MKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYV.GGEMFS
Sbjct: 61 MKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVPGGEMFS 120
Query: 117 HLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKR 176
HLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKR
Sbjct: 121 HLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKR 180
Query: 177 VKGRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKI 236
VKGRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKI
Sbjct: 181 VKGRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKI 240
Query: 237 VSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKV 296
VSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKV
Sbjct: 241 VSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKV 300
Query: 297 EAPFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF 336
EAPFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEF+EF
Sbjct: 301 EAPFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFSEF 340
>> 11
## Sbjct: ..\dat\PDB-sample\1q8wA.pdb
Protein length = 337 residues
Aligned residues = 336 residues
RMSD = 0.9435 Å
Seq identity = 97.32 % (327/336)
Seq similarity = 98.81 % (332/336)
Confidence-score = 0.9735
pC-value = 0.0387
TM-score, nQuery = 0.9813 (norm size: 336 res, d0: 6.69 Å)
Query: 1 -VKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMKI 59
VKEFLAKAKEDFLKKWE.P+QNTA:LDQF+RIKTLGTGSFGRVMLVKH.E+GNHYAMKI
Sbjct: 1 SVKEFLAKAKEDFLKKWENPAQNTAHLDQFERIKTLGTGSFGRVMLVKHMETGNHYAMKI 60
Query: 60 LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHLR 119
LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYV.GGEMFSHLR
Sbjct: 61 LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVPGGEMFSHLR 120
Query: 120 RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG 179
RIGRF.EPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG
Sbjct: 121 RIGRFXEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG 180
Query: 180 RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG 239
RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG
Sbjct: 181 RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG 240
Query: 240 KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP 299
KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP
Sbjct: 241 KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP 300
Query: 300 FIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF 336
FIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEF+EF
Sbjct: 301 FIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFSEF 337
>> 12
## Sbjct: ..\dat\PDB-sample\1rekA.pdb
Protein length = 338 residues
Aligned residues = 336 residues
RMSD = 1.0469 Å
Seq identity = 100.00 % (336/336)
Seq similarity = 100.00 % (336/336)
Confidence-score = 0.9735
pC-value = 0.0388
TM-score, nQuery = 0.9805 (norm size: 336 res, d0: 6.69 Å)
Query: 1 --VKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMK 58
VKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMK
Sbjct: 1 ESVKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMK 60
Query: 59 ILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHL 118
ILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHL
Sbjct: 61 ILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHL 120
Query: 119 RRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVK 178
RRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVK
Sbjct: 121 RRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVK 180
Query: 179 GRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVS 238
GRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVS
Sbjct: 181 GRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVS 240
Query: 239 GKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEA 298
GKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEA
Sbjct: 241 GKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEA 300
Query: 299 PFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF 336
PFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF
Sbjct: 301 PFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF 338
>> 13
## Sbjct: ..\dat\PDB-sample\1xhaA.pdb
Protein length = 350 residues
Aligned residues = 334 residues
RMSD = 0.7475 Å
Seq identity = 95.83 % (322/336)
Seq similarity = 98.21 % (330/336)
Confidence-score = 0.9695
pC-value = 0.0447
TM-score, nQuery = 0.9823 (norm size: 336 res, d0: 6.69 Å)
Query: 1 VK----------------EFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVM 44
EFLAKAKEDFLKKWE.P+QNTA:LDQF+RIKTLGTGSFGRVM
Sbjct: 1 --GNAAAAKKGSEQESVKEFLAKAKEDFLKKWENPAQNTAHLDQFERIKTLGTGSFGRVM 58
Query: 45 LVKHKESGNHYAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYM 104
LVKH.E+GNHYAMKILDKQKVVKLK+IEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYM
Sbjct: 59 LVKHMETGNHYAMKILDKQKVVKLKEIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYM 118
Query: 105 VMEYVAGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQG 164
VMEY..GGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENL+IDQQG
Sbjct: 119 VMEYAPGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLMIDQQG 178
Query: 165 YIQVTDFGFAKRVKGRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPF 224
YIQVTDFG:AKRVKGRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPF
Sbjct: 179 YIQVTDFGLAKRVKGRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPF 238
Query: 225 FADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFA 284
FADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFA
Sbjct: 239 FADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFA 298
Query: 285 TTDWIAIYQRKVEAPFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF 336
TTDWIAIYQRKVEAPFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEF+EF
Sbjct: 299 TTDWIAIYQRKVEAPFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFSEF 350
>> 14
## Sbjct: ..\dat\PDB-sample\3l9nA.pdb
Protein length = 334 residues
Aligned residues = 333 residues
RMSD = 1.0637 Å
Seq identity = 95.83 % (322/336)
Seq similarity = 98.21 % (330/336)
Confidence-score = 0.9670
pC-value = 0.0485
TM-score, nQuery = 0.9717 (norm size: 336 res, d0: 6.69 Å)
Query: 1 -VKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMKI 59
VKEFLAKAKEDFLKKWE+P+QNTA:LDQF+RIKTLGTGSFGRVMLVKHKE+GNHYAMKI
Sbjct: 1 SVKEFLAKAKEDFLKKWESPAQNTAHLDQFERIKTLGTGSFGRVMLVKHKETGNHYAMKI 60
Query: 60 LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHLR 119
LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEY..GGEMFSHLR
Sbjct: 61 LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYAPGGEMFSHLR 120
Query: 120 RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG 179
RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENL+IDQQGYI+VTDFGFAKRVKG
Sbjct: 121 RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLMIDQQGYIKVTDFGFAKRVKG 180
Query: 180 RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG 239
RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG
Sbjct: 181 RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG 240
Query: 240 KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP 299
KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP
Sbjct: 241 KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP 300
Query: 300 FIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF 336
FI .GPGDTSNFDDYEEEEIRVXINEKCGKEF+EF
Sbjct: 301 FI---PGPGDTSNFDDYEEEEIRVXINEKCGKEFSEF 334
>> 15
## Sbjct: ..\dat\PDB-sample\1cmkE.pdb
Protein length = 350 residues
Aligned residues = 335 residues
RMSD = 2.0660 Å
Seq identity = 96.43 % (324/336)
Seq similarity = 98.81 % (332/336)
Confidence-score = 0.9453
pC-value = 0.0812
TM-score, nQuery = 0.9303 (norm size: 336 res, d0: 6.69 Å)
Query: 1 VK---------------EFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVML 45
. EFLAKAKEDFLKKWE.P+QNTA:LDQF+RIKTLGTGSFGRVML
Sbjct: 1 G-NAAAAKKGSEQESVKEFLAKAKEDFLKKWENPAQNTAHLDQFERIKTLGTGSFGRVML 59
Query: 46 VKHKESGNHYAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMV 105
VKHKE+GNH+AMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLE+SFKDNSNLYMV
Sbjct: 60 VKHKETGNHFAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEYSFKDNSNLYMV 119
Query: 106 MEYVAGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGY 165
MEYV.GGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGY
Sbjct: 120 MEYVPGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGY 179
Query: 166 IQVTDFGFAKRVKGRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFF 225
IQVTDFGFAKRVKGRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFF
Sbjct: 180 IQVTDFGFAKRVKGRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFF 239
Query: 226 ADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFAT 285
ADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLK+GVNDIKNHKWFAT
Sbjct: 240 ADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKDGVNDIKNHKWFAT 299
Query: 286 TDWIAIYQRKVEAPFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF 336
TDWIAIYQRKVEAPFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEF+EF
Sbjct: 300 TDWIAIYQRKVEAPFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFSEF 350
>> 16
## Sbjct: ..\dat\PDB-sample\1sykA.pdb
Protein length = 350 residues
Aligned residues = 332 residues
RMSD = 2.7546 Å
Seq identity = 98.51 % (331/336)
Seq similarity = 98.81 % (332/336)
Confidence-score = 0.9213
pC-value = 0.1182
TM-score, nQuery = 0.8821 (norm size: 336 res, d0: 6.69 Å)
Query: 1 VK----------------EFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVM 44
EFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVM
Sbjct: 1 --GNAAAAKKGSEQESVKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVM 58
Query: 45 LVKHKESGNHYAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYM 104
LVKHKESGNHYAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYM
Sbjct: 59 LVKHKESGNHYAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYM 118
Query: 105 VMEYVAGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQG 164
VMEYVAGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQG
Sbjct: 119 VMEYVAGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQG 178
Query: 165 YIQVTDFGFAKRVKGRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPF 224
YIQVTDFGFAKRVKGRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIY+MAAGYPPF
Sbjct: 179 YIQVTDFGFAKRVKGRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYQMAAGYPPF 238
Query: 225 FADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFA 284
FADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFA
Sbjct: 239 FADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFA 298
Query: 285 TTDWIAIYQRKVEAPFIPKFKGPGD--TSNFDDYEEEEIRVXINEKCGKEFTEF 336
TTDWIAIYQRKVEAPFIPKFKGP TSNFDDYEEEEIRVXINEKCGKEFTEF
Sbjct: 299 TTDWIAIYQRKVEAPFIPKFKGP--GDTSNFDDYEEEEIRVXINEKCGKEFTEF 350
>> 17
## Sbjct: ..\dat\PDB-sample\3ag9A.pdb
Protein length = 325 residues
Aligned residues = 321 residues
RMSD = 2.5384 Å
Seq identity = 93.15 % (313/336)
Seq similarity = 94.64 % (318/336)
Confidence-score = 0.9096
pC-value = 0.1367
TM-score, nQuery = 0.8580 (norm size: 336 res, d0: 6.69 Å)
Query: 1 -VKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMKI 59
VKEFLAKAKEDFLKKWE.P+QNTA:LDQF+RIKTLGTGSFGRVMLVKH.E+GNHYAMKI
Sbjct: 1 SVKEFLAKAKEDFLKKWENPAQNTAHLDQFERIKTLGTGSFGRVMLVKHMETGNHYAMKI 60
Query: 60 LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHLR 119
LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYV.GGEMFSHLR
Sbjct: 61 LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVPGGEMFSHLR 120
Query: 120 RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG 179
RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG
Sbjct: 121 RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG 180
Query: 180 RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG 239
RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG
Sbjct: 181 RTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSG 240
Query: 240 KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP 299
KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP
Sbjct: 241 KVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEAP 300
Query: 300 FIPKFKGPGDTSNFDDYE---EEEIRVXINEKCGKEFTEF 336
FIP EEEIRVXINEKCGKEF+EF
Sbjct: 301 FIP---------------KFEEEEIRVXINEKCGKEFSEF 325
>> 18
## Sbjct: ..\dat\PDB-sample\4dfyA.pdb
Protein length = 311 residues
Aligned residues = 306 residues
RMSD = 3.2413 Å
Seq identity = 91.07 % (306/336)
Seq similarity = 91.07 % (306/336)
Confidence-score = 0.8600
pC-value = 0.2176
TM-score, nQuery = 0.7769 (norm size: 336 res, d0: 6.69 Å)
Query: 1 -VKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMKI 59
VKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMKI
Sbjct: 1 SVKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMKI 60
Query: 60 LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHLR 119
LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHLR
Sbjct: 61 LDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHLR 120
Query: 120 RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRVKG 179
RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGF
Sbjct: 121 RIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGF------ 174
Query: 180 RTWXLCG-TPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVS 238
TPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVS
Sbjct: 175 -------ATPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVS 227
Query: 239 GKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEA 298
GKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEA
Sbjct: 228 GKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKVEA 287
Query: 299 PFIPKFKGPGDTSNFDDYEE---EEIRVXINEKCGKEFTEF 336
PFI EEIRVXINEKCGKEFTEF
Sbjct: 288 PFI-----------------PKFEEIRVXINEKCGKEFTEF 311
>> 19
## Sbjct: ..\dat\PDB-sample\3e8dA.pdb
Protein length = 316 residues
Aligned residues = 298 residues
RMSD = 1.3609 Å
Seq identity = 38.10 % (128/336)
Seq similarity = 57.44 % (193/336)
Confidence-score = 0.8123
pC-value = 0.2999
TM-score, nQuery = 0.8577 (norm size: 336 res, d0: 6.69 Å)
Query: 1 VKEFLAKAKEDFLKKWETPSQNTAQLDQFDRIKTLGTGSFGRVMLVKHKESGNHYAMKIL 60
...++.FD.+K.LG.G+FG+V+LV+.K.+G.+YAMKIL
Sbjct: 1 ----------------------KVTMNDFDYLKLLGKGTFGKVILVREKATGRYYAMKIL 38
Query: 61 DKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMVMEYVAGGEMFSHLRR 120
.K+.++...++.HT:.E.R+LQ....PFL..L:++F+.:..L..VMEY..GGE+F.HL.R
Sbjct: 39 RKEVIIAKDEVAHTVTESRVLQNTRHPFLTALKYAFQTHDRLCFVMEYANGGELFFHLSR 98
Query: 121 IGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGYIQVTDFGFAKRV--- 177
...F+E..ARFY.A+IV..:EYLHS.D:+YRD+K.ENL++D+.G+I++TDFG:.K.
Sbjct: 99 ERVFTEERARFYGAEIVSALEYLHSRDVVYRDIKLENLMLDKDGHIKITDFGLCKE-GIS 157
Query: 178 -KGRTWXLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKI 236
.....X:CGTPEYLAPE++....Y.+AVDWW.LGV:+YEM..G..PF+.....:++E.I
Sbjct: 158 DGATMKXFCGTPEYLAPEVLEDNDYGRAVDWWGLGVVMYEMMCGRLPFYNQDHERLFELI 217
Query: 237 VSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWIAIYQRKV 296
:..:+RFP..:S.+.K.LL..LL+.D..+R:G...:...++..H+:F.+.+W..+.Q+K:
Sbjct: 218 LMEEIRFPRTLSPEAKSLLAGLLKKDPKQRLGGGPSDAKEVMEHRFFLSINWQDVVQKKL 277
Query: 297 EAPFIPKFKGPGDTSNFDDYEEEEIRVXINEKCG-------KEFTEF------- 336
..PF.P+.....DT..FDD ..F.+F
Sbjct: 278 LPPFKPQVTSEVDTRYFDD---------------EFTAQSITHFPQFDYSASIR 316
>> 20
## Sbjct: ..\dat\PDB-sample\7atvA.pdb
Protein length = 328 residues
Aligned residues = 257 residues
RMSD = 3.1805 Å
Seq identity = 17.86 % (60/336)
Seq similarity = 37.50 % (126/336)
Confidence-score = 0.6647
pC-value = 0.5892
TM-score, nQuery = 0.6660 (norm size: 336 res, d0: 6.69 Å)
Query: 1 VKEFLAKA--------------KED--FLKK-WETPSQNTAQLDQFDRIKTLGTGSFGRV 43
. +. +E....:....D.+..++.LG.G.+..V
Sbjct: 1 --------GSRARVYAEVNSLRS--REYW--DYEAHVPSWGNQDDYQLVRKLGRGKYSEV 48
Query: 44 MLVKHKESGNHYAMKILDKQKVVKLKQIEHTLNEKRILQ-AVNFPFLVKLEFSFKDNS-- 100
....:..:.....:KIL.. .+.:....E.+IL+ ......++KL..+.KD..
Sbjct: 49 FEAINITNNERVVVKILKP------VKKKKIKREVKILENLRGGTNIIKLIDTVKDPVSK 102
Query: 101 NLYMVMEYVAGGEMFSHL--RRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENL 158
...+V.EY+...+ :....:++...RFY..++:..:+Y.HS..+++RD:KP.N:
Sbjct: 103 TPALVFEYINNTD-----FKQLYQILTDFDIRFYMYELLKALDYCHSKGIMHRDVKPHNV 157
Query: 159 LID-QQGYIQVTDFGFAKRVKG-RTWXLC-GTPEYLAPEIILS-KGYNKAVDWWALGVLI 214
+ID QQ..+::.D:G:A:.... :.+.:. .+..+..PE++:. +.Y+.+:D.W+LG.++
Sbjct: 158 MIDHQQKKLRLIDWGLAEFYHPAQEYNVRVASRYFKGPELLVDYQMYDYSLDMWSLGCML 217
Query: 215 YEMAAGYPPFF-ADQPIQIYEKIVS------------------------------GKVRF 243
..M.....PFF .........+I. .+.R
Sbjct: 218 ASMIFRREPFFHGQDNYDQLVRIA-KVLGTEELYGYLKKYHIDLDPHFNDILGQHSRKR- 275
Query: 244 ---------PSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFATTDWI-AIYQ 293
....S.+..DLL..LL:.D..+R ....+...H.+F + .+
Sbjct: 276 WENFIHSENRHLVSPEALDLLDKLLRYDHQQR-----LTAKEAMEHPYF----Y-PVV-- 323
Query: 294 --RKVEAPFIPKFKGPGDTSNFDDYEEEEIRVXINEKCGKEFTEF-- 336
:
Sbjct: 324 KEQ------------------------------------------SQ 328
>> 21
## Sbjct: ..\dat\PDB-sample\3at2A.pdb
Protein length = 334 residues
Aligned residues = 239 residues
RMSD = 2.8027 Å
Seq identity = 16.67 % (56/336)
Seq similarity = 36.01 % (121/336)
Confidence-score = 0.6530
pC-value = 0.6148
TM-score, nQuery = 0.6360 (norm size: 336 res, d0: 6.69 Å)
Query: 1 VKEFLAKAKED---------------------FLKK--WE-TPSQNTAQLDQFDRIKTLG 36
+ + +........D.+..++.LG
Sbjct: 1 -----------SGPVPSRARVYTDVNTHRPREY---WDY-ESHVVEWGNQDDYQLVRKLG 45
Query: 37 TGSFGRVMLVKHKESGNHYAMKILDKQKVVKLKQIEHTLNEKRILQ-AVNFPFLVKLEFS 95
.G.+..V....:..:.....:KIL.. .+.:....E.+IL+ ....P.++.L...
Sbjct: 46 RGKYSEVFEAINITNNEKVVVKILKP------VKKKKIKREIKILENLRGGPNIITLADI 99
Query: 96 FKD--NSNLYMVMEYVAGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDL 153
.KD :....+V.E+V...+. .:L.:.. ++...RFY..+I:..:+Y.HS+.+++RD:
Sbjct: 100 VKDPVSRTPALVFEHVNNTDF-KQLYQTL--TDYDIRFYMYEILKALDYCHSMGIMHRDV 156
Query: 154 KPENLLIDQ--QGYIQVTDFGFAKRVKG-RTWXLC-GTPEYLAPEIILS-KGYNKAVDWW 208
KP.N:+ID :..+::.D:G:A:.... :.+.:. .+..+..PE++:. +.Y+.+:D.W
Sbjct: 157 KPHNVMID-HEHRKLRLIDWGLAEFYHPGQEYNVRVASRYFKGPELLVDYQMYDYSLDMW 215
Query: 209 ALGVLIYEMAAGYPPFFADQPIQIYEKIVSGKVRFPS----------------------- 245
+LG.++..M.....PFF
Sbjct: 216 SLGCMLASMIFRKEPFF--------------------HGHDNYDQLVRIAKVLGTEDLYD 255
Query: 246 -----------------------------------HFSSDLKDLLRNLLQVDLTKRFGNL 270
..S.+..D:L..LL:.D...R
Sbjct: 256 YIDKYNIELDPRFNDILGRHSRKRWERFVHSENQHLVSPEALDFLDKLLRYDHQSR---- 311
Query: 271 KNGVNDIKNHKWFATTDWIAIY-QRKVEAPFIPKFKGPGDTSNFDDYEEEEIRVXINEKC 329
....+...H.+F +..+ .:
Sbjct: 312 -LTAREAMEHPYF----YTVV-KDQ----------------------------------- 330
Query: 330 GKEFTEF---- 336
Sbjct: 331 -------ARMG 334formatdb
formatdb
on Windows
.\formatdb.exe "..\dat\PDB-sample\*.pdb" -db mySarstDbon Linux/macOS
./formatdb "../dat/PDB-sample/*.pdb" -db mySarstDbExample result
D:\software>cd SARST2-v2.0.30-Win10+.x86_64\bin
D:\software\SARST2-v2.0.30-Win10+.x86_64\bin>.\formatdb.exe "..\dat\PDB-sample\*.pdb" -db mySarstDb
+------------------------------------------------------------------------------+
| ____ _ ____ ____ _____ ____ |
| / ___| / \ | _ \/ ___|_ _|___ \ Structural similarity search |
| \___ \ / _ \ | |_) |___ \ | | __) | Aided by |
| ___) | ___ \| / ___) || | / __/ Ramachandran Sequential Transformation |
| |____/_/ \_\_|\_\|____/ |_| |_____| (v2.0.30, build 20250609) |
|______________________________________________________________________________|
| |
| Please cite our paper(s) should you find this software helpful, |
| Wei-Cheng Lo, et al. Nature Communications 2025, 10.1038/s41467-025-63757-9 |
| Wei-Cheng Lo, et al. Nucleic Acids Research 2009, 10.1093/nar/gkp291 |
| Wei-Cheng Lo, et al. BMC Bioinformatics 2007, 10.1186/1471-2105-8-307 |
+------------------------------------------------------------------------------+
# Processed # Queued === Structure files ===
100 100 In queue : ..\dat\PDB-sample\*.pdb
Subjects: ..\dat\PDB-sample\*.pdb
Dataset size = 100 structure(s)To check the contents of current directory, you can use the following command:
on Windows
diron Linux/macOS
lsIf the program run successfully, you will see 7 files with the file stem mySarstDb, along with a file named mySarstDb that records the execution time.
readdb
readdb (a database formatted by formatdb is required)
on Windows
.\readdb.exe mySarstDb seqs.fastaon Linux/macOS
./readdb mySarstDb seqs.fastaExample result
D:\software>cd SARST2-v2.0.30-Win10+.x86_64\bin
D:\LabWork\working\250715_test\SARST2-v2.0.30-Win10+.x86_64\bin>.\readdb.exe mySarstDb seqs.fasta
+------------------------------------------------------------------------------+
| ____ _ ____ ____ _____ ____ |
| / ___| / \ | _ \/ ___|_ _|___ \ Structural similarity search |
| \___ \ / _ \ | |_) |___ \ | | __) | Aided by |
| ___) | ___ \| / ___) || | / __/ Ramachandran Sequential Transformation |
| |____/_/ \_\_|\_\|____/ |_| |_____| (v2.0.30, build 20250609) |
|______________________________________________________________________________|
| |
| Please cite our paper(s) should you find this software helpful, |
| Wei-Cheng Lo, et al. Nature Communications 2025, 10.1038/s41467-025-63757-9 |
| Wei-Cheng Lo, et al. Nucleic Acids Research 2009, 10.1093/nar/gkp291 |
| Wei-Cheng Lo, et al. BMC Bioinformatics 2007, 10.1186/1471-2105-8-307 |
+------------------------------------------------------------------------------+
Output type: Amino acid sequence
## Retrieving the entry list... (mySarstDb)
=> 100 entries
## Writing sequences...
100
## Finished
=> 100 sequences written to 'seqs.fasta'.To view the contents of seqs.fasta, you can use the following command:
on Windows
type seqs.fastaon Linux/macOS
cat seqs.fastaHow to exit when done?
You can use the command
exit